Breast Cancer Metabolism
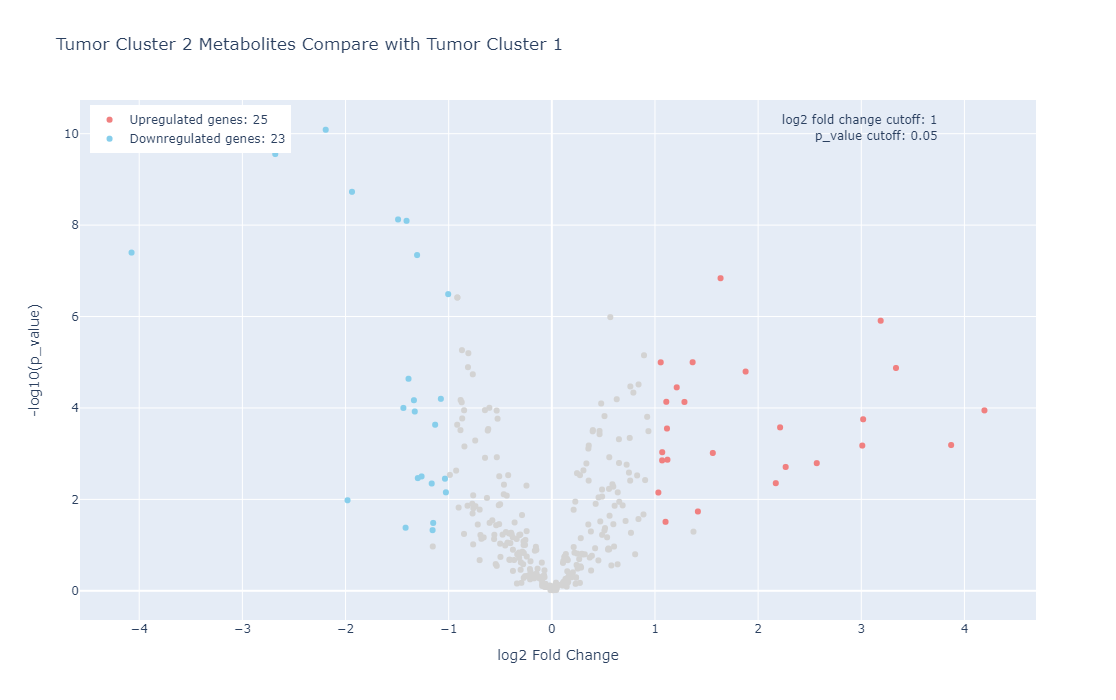
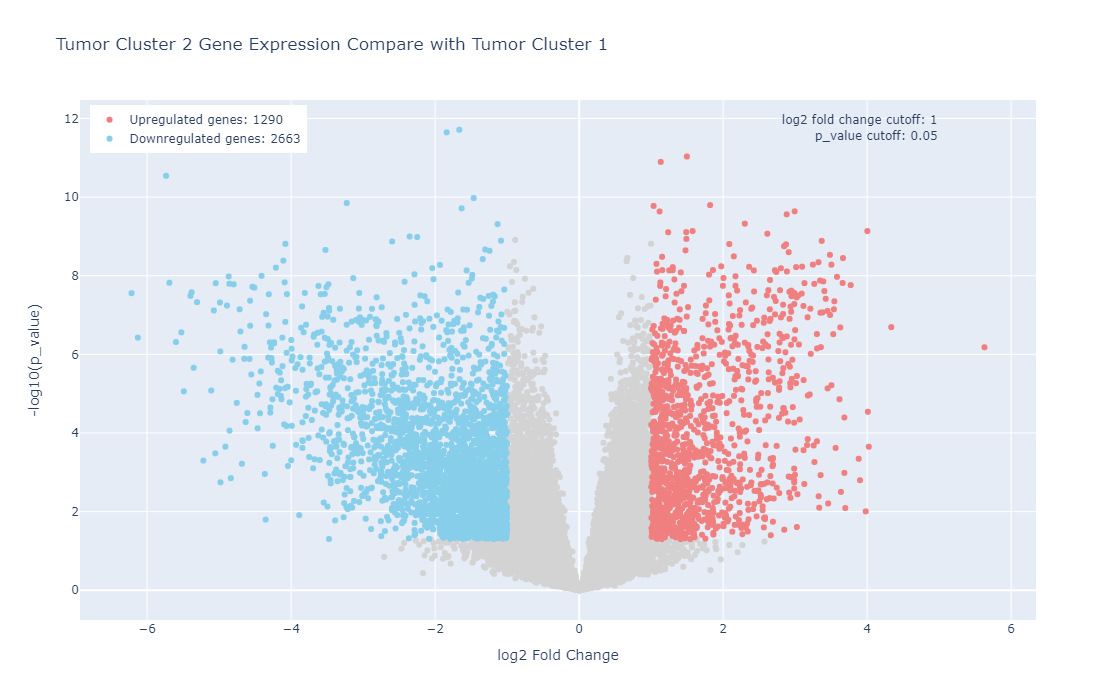
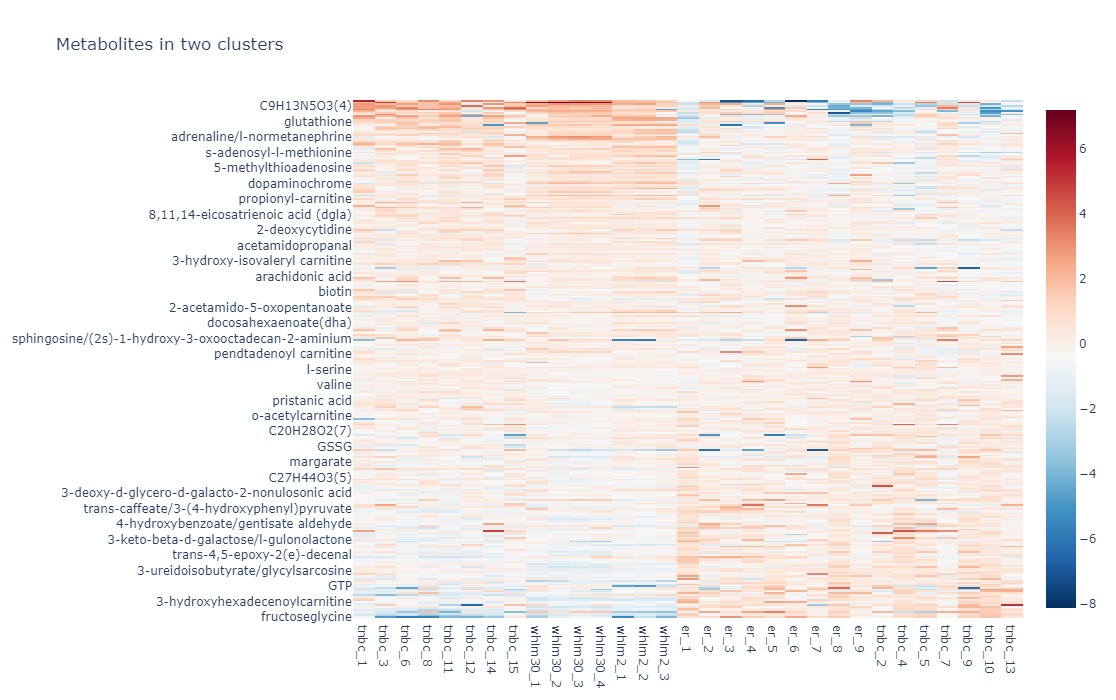
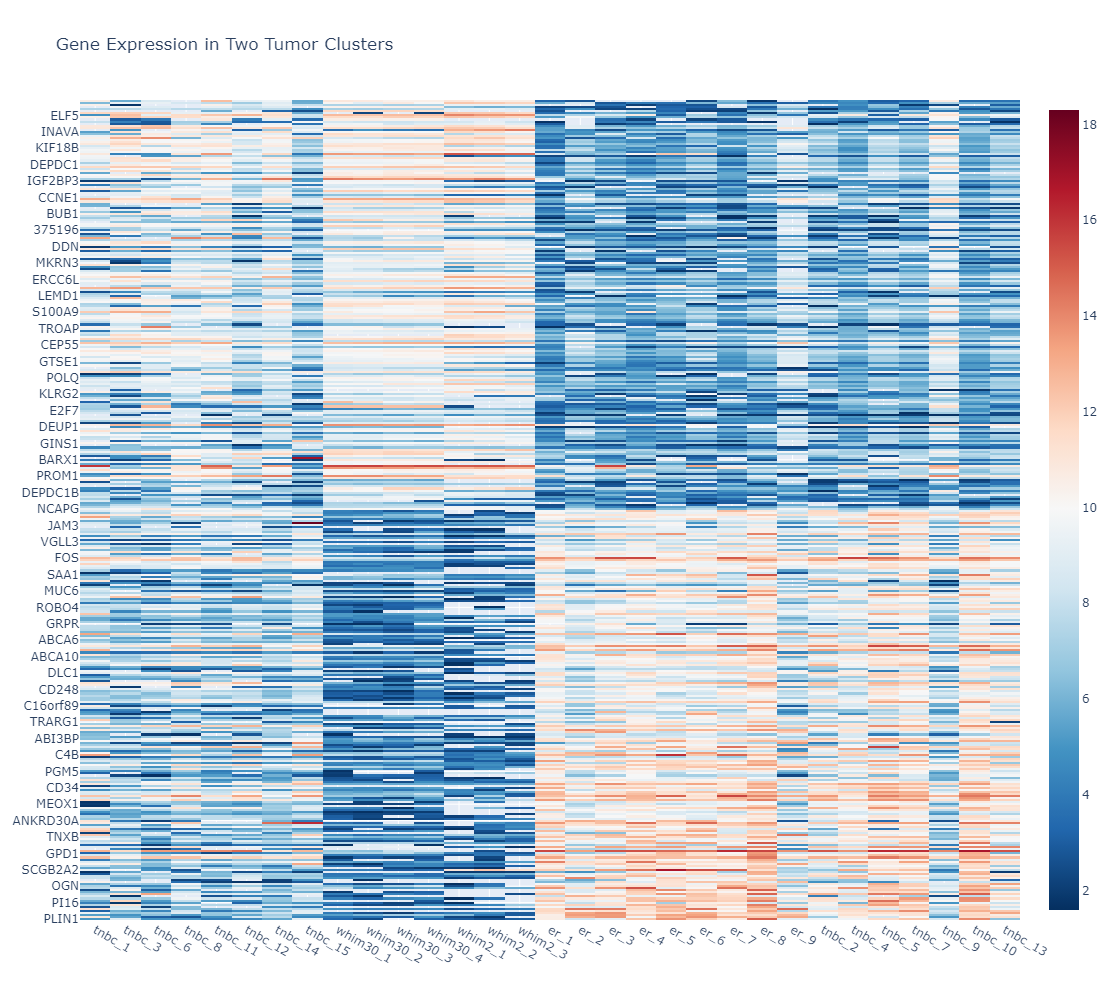
Metabolic dysregulation is one of the distinctive features in breast cancer. However, examining the metabolic features in various subtypes of breast cancer in their relationship to gene expression features in a physiologically relevant setting remains understudied. In this project, we performed metabolic profiling on 9 pairs of normal breast tissues and ERα+ breast tumors, 15 Triple-Negative Breast Cancer (TNBC) tumors, two models of TNBC patient-derived xenografts (PDXs) (WHIM2 and WHIM30). We extracted metabolites from these tumors followed by a liquid chromatography, high-resolution mass spectrometry (LC-MS) analysis. we next performed a hierarchical cluster analysis using the quantitative metabolic data coming from these samples and our unsupervised cluster divided these patients into two distinctive clusters (Cluster 1 and Cluster 2). We also performed RNA-Seq for most of these samples. In addition, we also examined metabolic features of 4 ERα+ breast cancer cell lines as well as 4 TNBC cell lines grown on petri dishes or grown as tumors in mice. We provide these data here so that the community can make use of the data for their research needs.